In this post I’ll be using R to run Principal Component Analysis on the same dataset used on this which is part of the Udacity Data Analyst Nanodegree. The process I’ll follow is pretty much the same as the one on the webcast but I hope to be adding some extra information to it. I also would like to than Udacity, Sheng Kung and Nikolay for the amazing webcast and especially for the detailed answers to my questions on the forums.
1) What is PCA and what is it good for?
· Very efficient and automatic way of combining features in order to simplify a very large and complicated dataset;
· If we have 2 features, we want to make sure that they are as independent as possible from each other; that each one contains the “purest” kind of information on their own;
· It does that by trying to identify correlation between features by considering a new feature that is some sort of hybrid between others, to make the model simpler and easier to fit on the data;
· One down side is that PCA are vectors over the initial feature so they can be harder to interpret than the initial features;
· One of the assumptions on PCA is that the data is continuous and that it follows roughly a normal distribution. There are alternate techniques that handle other types of data;
2) Data
As I said, I’ll be using the same dataset used on the website but to make the process easier I skipped the data cleaning process and am loading a “ready-to-go” dataset. You can find details about the dataset and the cleaning process on this ipython notebook. (I downloaded it from box 15 on the notebook).
The data set is pretty simple, it contains information about 105 countries around the globe which will all be considered as features for the sake of the PCA example.
df <- read.csv ("cleaned.csv")
head(df)
|
Country |
completion_male |
completion_female |
income_per_person |
employment |
life_expectancy |
|
Argentina |
97.91143 |
102.10666 |
9.465967 |
58.4 |
75.5 |
|
Armenia |
91.53127 |
94.58897 |
8.749288 |
39.4 |
72.2 |
|
Austria |
100.63802 |
101.12819 |
10.656714 |
57.1 |
80 |
|
Azerbaijan |
91.68856 |
90.66259 |
9.264091 |
59.3 |
69.9 |
|
Bahamas |
99.76959 |
96.84352 |
10.133238 |
66.9 |
72.1 |
3) Running PCA:
PCA can be run using the prcomp command. The command offers the possibility of normalizing the data using the “center” and “scale” parameters (more info about normalizing can be found here). The reason you should always normalize the data is, imagine that you are trying to run PCA on 2 features A and B that range from, let’s say, A: 1 and 100 and B: 0 and 10. If you do that without normalizing the data, PCA is going to find a large principal component on the A variable so it is going to follow it rather than trying to find any correlation between the features.
Here I’m calling the prcomp command with center and scale = TRUE (If you omit these parameters you’ll see that the values outputted (weights) will be quite different):
prcomp(df[,2:6],center = TRUE,scale = TRUE)
Standard deviations:
[1] 1.8207722 0.9510142 0.7172067 0.4642723 0.2245580
Rotation:
PC1 PC2 PC3 PC4 PC5
completion_male -0.5028374 -0.01732469 0.5155280 0.05640884 -0.691305540
completion_female -0.5088654 -0.01884488 0.4716004 -0.06446605 0.717034253
income_per_person -0.4686439 0.23200303 -0.4755441 -0.70319643 -0.076942116
employment 0.2264034 0.93671022 0.2659600 -0.02261103 0.008335095
life_expectancy -0.4661813 0.26094689 -0.4636238 0.70545424 0.044373047
We’ll be looking at these values in a moment but to exemplify what the normalization is doing, I will manually normalise the initial dataset by subtracting each one of its value by the feature’s mean and dividing by its standard deviation:
df$completion_male <- ((df$completion_male - mean(df$completion_male)) / sd(df$completion_male)) df$completion_female <- ((df$completion_female - mean(df$completion_female)) / sd(df$completion_female)) df$income_per_person <- ((df$income_per_person - mean(df$income_per_person)) / sd(df$income_per_person)) df$employment <- ((df$employment - mean(df$employment)) / sd(df$employment)) df$life_expectancy <- ((df$life_expectancy - mean(df$life_expectancy)) / sd(df$life_expectancy)) head(df)
Here’s how the Normalized data looks like:
|
Country |
completion_male |
completion_female |
income_per_person |
employment |
life_expectancy |
|
Argentina |
0.500062073 |
0.685201535 |
0.30719635 |
0.016051348 |
0.63137587 |
|
Armenia |
0.149531869 |
0.339307131 |
-0.272843386 |
-1.86760039 |
0.26952447 |
|
Austria |
0.649862727 |
0.640181416 |
1.270919761 |
-0.11283039 |
1.1248096 |
|
Azerbaijan |
0.158173486 |
0.158651521 |
0.143808985 |
0.105276729 |
0.01732501 |
|
Bahamas |
0.60215061 |
0.443040622 |
0.847247396 |
0.85873765 |
0.25855927 |
Now, if I run PCA with the center and scale parameters = FALSE on the normalized data, you can see that I get the same result as the previous command.
pca<- prcomp(df[,2:6],center = FALSE,scale = FALSE)
pca
Standard deviations:
[1] 1.8207722 0.9510142 0.7172067 0.4642723 0.2245580
Rotation:
PC1 PC2 PC3 PC4 PC5
completion_male -0.5028374 -0.01732469 0.5155280 0.05640884 -0.691305540
completion_female -0.5088654 -0.01884488 0.4716004 -0.06446605 0.717034253
income_per_person -0.4686439 0.23200303 -0.4755441 -0.70319643 -0.076942116
employment 0.2264034 0.93671022 0.2659600 -0.02261103 0.008335095
life_expectancy -0.4661813 0.26094689 -0.4636238 0.70545424 0.044373047
4) Principal Components:
Each component corresponds to the coefficients (weights) to be applied to each feature (the data points) in order to get to the new resulting point. PCA can come up with a number of orthogonal components that is equal to the number of features we fit, so if we have 5 features on the model, the maximum number of principal components is 5. By running a summary on the pca object, we can see that the first PCA explain 66% of the data’s variance, the second 18% and so on:
summary(pca)
Importance of components:
PC1 PC2 PC3 PC4 PC5
Standard deviation 1.821 0.9510 0.7172 0.46427 0.22456
Proportion of Variance 0.663 0.1809 0.1029 0.04311 0.01009
Cumulative Proportion 0.663 0.8439 0.9468 0.98991 1.00000
The prcomp function offers a “tol” parameter that can be used to limit the number of component to be returned. The value indicates the magnitude below which components should be omitted. (Components are omitted if their standard deviations are less than or equal to “tol” times the standard deviation of the first component.) . For example, had we called the function above with tol =0.4, we’d have gotten only 2 principal components because 1.821 * 0.5 = 0.9105 and PC3’s standard deviation is 0.7172
The principal components can be found by running pca[2] which will output a matrix containing the features as rows and the principal components as columns:
pca[2] #as.data.frame(components$rotation)
|
row.names |
PC1 |
PC2 |
PC3 |
PC4 |
PC5 |
|
completion_male |
-0.5028374 |
-0.01732469 |
0.515528 |
0.05640884 |
-0.69130554 |
|
completion_female |
-0.5088654 |
-0.01884488 |
0.4716004 |
-0.06446605 |
0.717034253 |
|
income_per_person |
-0.4686439 |
0.23200303 |
-0.4755441 |
-0.70319643 |
-0.07694212 |
|
employment |
0.2264034 |
0.93671022 |
0.26596 |
-0.02261103 |
0.008335095 |
|
life_expectancy |
-0.4661813 |
0.26094689 |
-0.4636238 |
0.70545424 |
0.044373047 |
Note that all 5 principal components are generated from all five features (principal components are a linear combination over all features). If we had requested prcomp to generate a different number or pcs the order of them would be the same since the data isn’t actually changing. For example, PC1 will always be (-0.5028374, -0.5088654 and so on…) since those vectors should still be the directions of maximal variance on the five features we are investigating.
Even though each principal component is generated from all five features, one thing that you might find from your principal component transformation is that there may be certain features that are highly correlated with one another and you might see a principal component that has a very large weight on a handful of features.
Features with similar coefficients in the same direction (both positive or both negative) are obviously similar, but features of the same magnitude but opposite directions are also interesting for their negative correlations. In terms of overall magnitude, coefficients do not need to all be large, only similar.
The overall length of the principal component vector is 1 (the sum of each weight squared), so if we observe a coefficient that has a large magnitude, then we know that most of the weight of that principal component is on that single feature and the rest of the weights will be relatively small. You’ll tend to see correlated features have smaller coefficient values, due to the need to ‘share’ weight on each of the features.
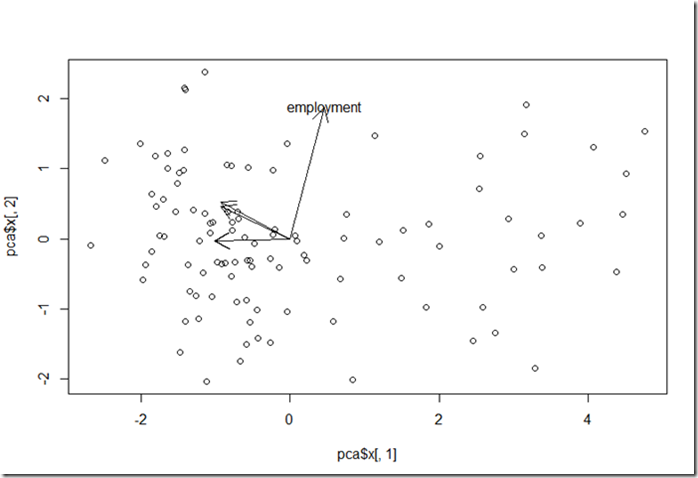
In this case, we can see that the second principal component represents the variation contributed by the “employment” feature.
Another interesting feature of the pca object is X (fifth feature on the pca list), which contains the transformed data (the original data with PCA weights applied):
as.data.frame(pca$x[1:5,])
|
PC1 |
PC2 |
PC3 |
PC4 |
PC5 |
|
-1.0348 |
0.2295 |
0.1464 |
0.2131 |
0.1501 |
|
-0.6685 |
-1.7514 |
-0.2548 |
0.4108 |
0.1573 |
|
-1.7981 |
0.4594 |
-0.5189 |
-0.1023 |
-0.0390 |
|
-0.2119 |
0.1308 |
0.1079 |
-0.0926 |
-0.0050 |
|
-0.8514 |
1.0496 |
0.2250 |
-0.4274 |
-0.1452 |
So, for example, if we get the first row of our ds dataset, Argentina, (remember that it contains the normalized values because I did it manually) and do the matrix multiplication between its values and the first principal component, we’ll get cell pca$x[1,1]:
|
|
0.500062073 * –0.5028374 +
0.685201535 * –0.5088654 +
0.30719635 * –0.4686439 +
0.016051348 * 0.2264034 +
0.63137587 * –0.4661813 = –1.0348
5) PCA’s Properties:
The resulting components from PCA are independent from each other. The information contained in one doesn’t tell anything about the value that is on the other features. So when we fit it to a model, each of these features stands on its own by telling something different between the relationship between our predictors and out outcome. Also, the components are orthogonal to each other and normalized to have length 1:
components <- pca[2]
#Components are orthogonal to each other (dot product = 0)
components$rotation[,3] %*% components$rotation[,1]
[,1]
[1,] 1.387779e-16
#Components are normaled to have length 1.
sum(components$rotation[,1]**2)
[1] 1
6) Plotting:
If we plot this in the principal component space (where each dot represents a country plotted with its 2 PCAs – let’s use PC1 and PC2), with an arrow pointing from the origin to the point implied by the row’s coordinates, we can see how well each feature aligns with each principal component. If we see an arrow aligned strongly with one of the axes, then we can say that the principal component on that axis may represent the feature fairly well. (For example, the fourth row has a very strong alignment with the second principal component.)
We can also see, if multiple vectors point in the same direction, how much similarity there is between features. (This can be seen in the first and second rows of the array, as well as the third and fifth rows.)
pca1<-components$rotation[,1]
pca2<-components$rotation[,2]
plot(pca$x[,1], pca$x[,2])
arrow_size=2
arrows(0,0,pca1[1]* arrow_size,pca2[1]* arrow_size)
arrows(0,0,pca1[2]* arrow_size,pca2[2]* arrow_size)
arrows(0,0,pca1[3]* arrow_size,pca2[3]* arrow_size)
arrows(0,0,pca1[4]* arrow_size,pca2[4]* arrow_size)
text(pca1[4]* arrow_size,pca2[4]* arrow_size, row.names(pca$rotation)[4])
arrows(0,0,pca1[5]* arrow_size,pca2[5]* arrow_size)
|
|
completion_male |
completion_female |
income_per_person |
employment |
life_expectancy |
|
PC1 |
-0.50283735 |
-0.50886538 |
-0.4686439 |
0.2264034 |
-0.4661813 |
|
PC2 |
-0.01732469 |
-0.01884488 |
0.232003 |
0.9367102 |
0.2609469 |
7) Extra points on Interpreting the results:
The explained variance on each principal component can be very useful on the interpretation. Since there is a good amount of variance explained by both the first and second principal component, we can take a look at both of these components together when trying to see if there are features that go well with one another. If the first component had a higher amount of explained variance and the second component a smaller amount of explained variance, then we might well believe that there is only one major signal in the data that is covered by all of the components except for the fourth. With the data as it is, the second component is important enough that we might be better-suited to consider the first and second features as one pair of related features, and the third and fifth as a second pair.
If we see this behaviour in our principal components, then we can actually go back and create hybrid features and then substitute them in for our original features in the original feature space. When it comes to creating composite features based on the results of PCA, it certainly helps to have domain knowledge. And if you don’t have that knowledge, it is a good opportunity to do some additional exploration and research to see if you can understand the underlying relationship between features. If you have documentation for your dataset, look there; performing additional visualizations can also be a big help to understanding the data. Data analysis is a process where you do need to move between steps in an iterative fashion, from wrangling to exploration to modelling.
As for selecting a good number of principal components to reduce down to, you can look at the amount of variance explained by each component. If you start out with a large number of components, then you can see the importance of each component on explaining variation in the original data. Once the values get very small, you can make an assumption that the components are beginning to fit to the random noise in the data rather than the true signal. If you look at the size of the explained variance values or the trend in the cumulative sum of explained variance coefficients (visualizing these in plots will be quite useful), this can help you make a judgement on how many components you want to include in your algorithm.
Interpretation of the output of principal component analysis is quite tricky since there are a lot of moving parts and principal components may not always be easily interpretable. I don’t think there’s any hard, deterministic rules to follow; one of the most important skills that you can have is to be able to synthesize multiple bits of information together and make your own conclusions.
However, it’s also important to realize in practice, it may not be possible to see such clean correlations and relationships between features. If we have a lot of features, it may be difficult to find a coherent interpretation for each principal component. This is one of the downsides of PCA that it may be a good dimensionality reduction technique, but it certainly has the possibility of returning un-interpretable components.